In my previous post, I have explained multicollinearity and its sources, effect on inference and method of detecting the presences of multicollinearity. Presence of multicollinearity in data may dramatically impact the usefulness of Regression Model. So, now I am going to explain a method for dealing with multicollinearity in regression analysis which is Principal-Components Regression(PCR) . Let's get start it.
Introduction :
Principal component regression (Massy, 1965; Jolliffe, 1982) is a widely used two-stage procedure:
- First performs Principal Component Analysis(PCA) (Pearson,1901; Jolliffe, 2002).
- Next considers are Regression Model in which the selected principal components are regarded as new regressors.
However, we should remark that PCA is based only on the regressors, so the principal components are not selected using the information on the response variable. PCR is most popular technique for analyzing multiple regression data that suffer from multicollinearity. PCR is also sometimes used for general dimension reduction.
Assumption :
- Linearity of the phenomenon measured.
- Constant variance of the error terms.
- Independence of the error terms.
- Normality of the error term distribution.
Details of the method :
Data representation:
Following centering the standard Gauss-markov linear regression model for y on X can be represented as,
Where,
Each of the 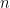 rows of
rows of  denotes one set of observation for the
denotes one set of observation for the  dimensional predictor variables and the respective entry of
dimensional predictor variables and the respective entry of 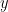 denotes the corresponding response variable.
denotes the corresponding response variable.
Consider the canonical form of the model
Where,
are referred to as principal components.
Fitting of PCR for the Acetylene Data using Python :
I am going to use same dataset for Regression Analysis which I have used in my Multicollinearity : Introduction, Sources, Effect and Diagnostics blog
Example : The acetylene Data
 |
Table 1 |
Table 1 present data concerning the percentage of conversion of 'n-heptane to acetylene' and three explanatory variables(predictor variables or predictors) 'Reactor Temperature (oC)', 'Ratio of H2 to n-Heptane ( mole ratio)' and 'Contact Time (sec)'.
Step 1: Performs Principal Component Analysis(PCA) on explanatory variable
Import libraries and read data from csv
Check correlation between explanatory variables
Here, we can see that correlation present between explanatory variables. So let's do further regression analysis using PCA. Before calculating PCA, we need to bring all explanatory variables into same unit as they are having different unit. I am going to use StandardScaler() for scaling.
Explained Variance : The explained variance tells you how much information (variance) can be attributed to each of the principal components. This is important as while you can covert high dimensional space to two dimensional space (Data Reduction), you lose some of the information when you do this.
Now, we are having three principal component as having three explanatory variables. You can see that the first principal component contains 68.66% of the variance and second principal component contains 29.95% of the variance. Together, the two components contain 98.61% of the information. Therefore, I am going to used first two principal components which are contain maximum information of explanatory variables. We can select any number of principal components according to your data, features and variance that explained by PCA, such that we can achieve high accuracy through model which is good for prediction.
Step 2: Regression Model (selected principal components are regarded as new regressors)
Here, we can see that coefficient of determination is 0.90616 which is good accuracy measure in regression analysis. Coefficient of determination telling us that, how much variation is explaining this model.
Conclusion :
Our PCR model can explaining 90.61% variability in the whole data using first two principal components, just 9.39% information is unexplainable in this model, there might be some error's contribution in unexplainable information as accuracy never ever be 100% in statistics (error always be there), which is good for prediction.
Advantages:
- PCR can perform regression when the explanatory variables are highly correlated or even collinear.
- PCR is intuitive : We replace the basis
with an orthogonal basis of principal components, drop the components that do not explain much variance, and regress the response onto the remaining components.
- PCR is automatic : The only decision you need to make is how many principal components to keep
- We can run PCR when there are more variables than observations(wide data)
Disadvantages :
- PCR does not consider the response variable when deciding which principal components to drop. The decision to drop components is based only on the magnitude of the variance of the components.
- There is no a priori reason to believe that the principle components that best predict the response
Endnote :
PCR is a technique for computing regression when the explanatory variable are highly correlated. If has several advantages, but the main drawback of PCR is that the decision about how many principal to keep does not depend on the response variable. Consequently, some of the variables that you keep might not be strong predictors of the response, and some of the components that you drop might be excellent predictors. A good alternatives are Ridge Regression and Partial Least Squares(PLS) Regression.
---Thank you---














